 Choose language
Choose language
< Return to main menu
 Choose language
Choose language

In chapters 13 and 14, we discussed how torsional strain controls the selectivity observed in peracid epoxidation of norbornene, and the application of torsion scan to assess whether compounds of interest could exhibit atropisomerism, respectively. In this chapter, we’ll discuss further the application of QM torsion scan to guide drug discoveries of atropisomers.
Atropisomers are stereoisomers arising because of hindered rotation about a single bond. This could be exemplified with the biphenyl structure shown in Figure 1. The four substituents A, A’, B, and B’ on both sides of the single bond of the biphenyl analog exert steric hindrance for rotation. When the energy barrier of rotation is large enough, the pair of atropisomers cannot rapidly interconvert at room temperature and could be separated by chiral chromatography.
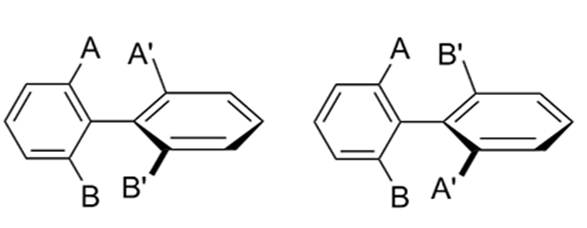
Figure 1. Atropisomer [1]
According to statistics, 80% of kinase inhibitors approved by FDA have axial chirality [2]. It was reported in the literature [3] that axial chiral molecules could have different half-lives (T1/2) for racemization, depending on the difference in energy barriers for rotation. Atropisomerism can be divided into three classes.
Class 1: DERot <20 kcal/mol, half-life (T1/2) in seconds. Atropisomers can rotate freely and quickly, behaving as single compounds.
Class 2: 20 kcal/mol < DERot < 30 kcal/mol, T1/2 between hours and days, separable with chiral SFC.
Class 3: DERot > 30 kcal/mol, T1/2 in years, the atropisomer is very stable, separable with chiral SFC, could be developed as a single compound.
It has been demonstrated that QM torsion scan analyses could be used to evaluate the energy barrier of the axial rotation, estimate their interconversion rate, guide us to increase or decrease the energy barriers in target designs, provide guidance whether chiral SFC separations are necessary, shorten cycle time during lead optimization, and adopt proper development strategies [4].
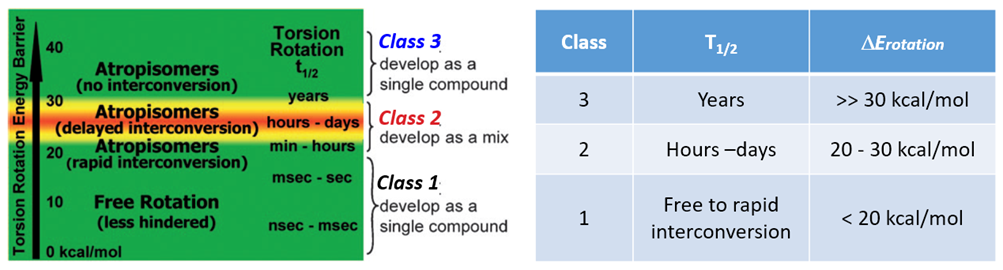
Figure 2. Classification of Atropisomers [3]
Next, we’ll use Afloqualone to illustrate the use of QM torsion scan to estimate energy barriers of atropisomer interconversion. Shown in Figure 3 is the overall procedure. First, the low-energy conformation of Afloqualone was calculated using MMFF (Spartan’20). Then from the menu, choose Geometry, select Measure Dihedral, click on the C1-N2-C3-C4 atoms in order, and select the dihedral angle to rotate. The torsion scan was then set to calculate from a dihedral angle of 15º to 345º, with an increment of 15º, in 23 steps (excluding the conformers with dihedral angle of 0º to 360º which will have the methyl and fluoromethyl group crashing into one another) using Hartree-Fock 6-31G**method (gas phase) [3].
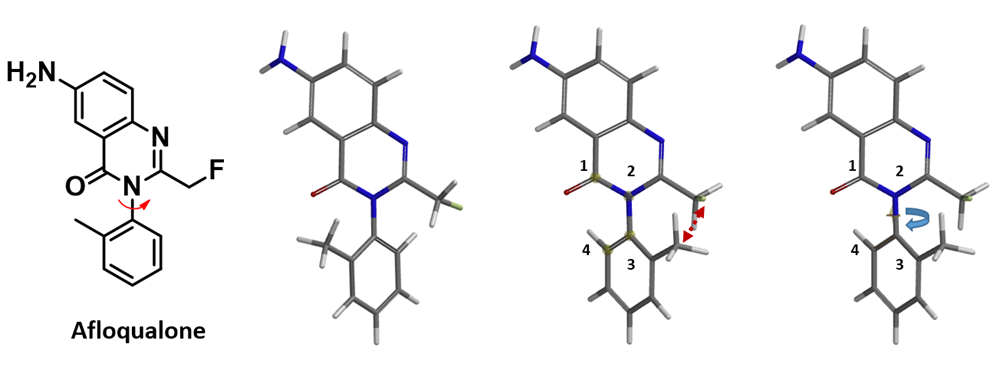
Figure 3. Torsion scan calculation procedure as exemplified with Afloqualone [4]
The calculation results are shown in Figure 4. The two lowest energy conformations have dihedral angles of 90º and 270º, respectively, both with a relative energy of 0 kcal/mol. Their interconversion will require the molecules to go through the conformation with an 180º dihedral angle, with the methyl and the carbonyl group coplanar to one another, and an energy barrier of 34.96 kcal/mol. As such, Afloqualone belongs to class 3 atropisomers, separable with chiral SFC, having no significant interconversion at room temperature, and can be developed as single chiral compounds.
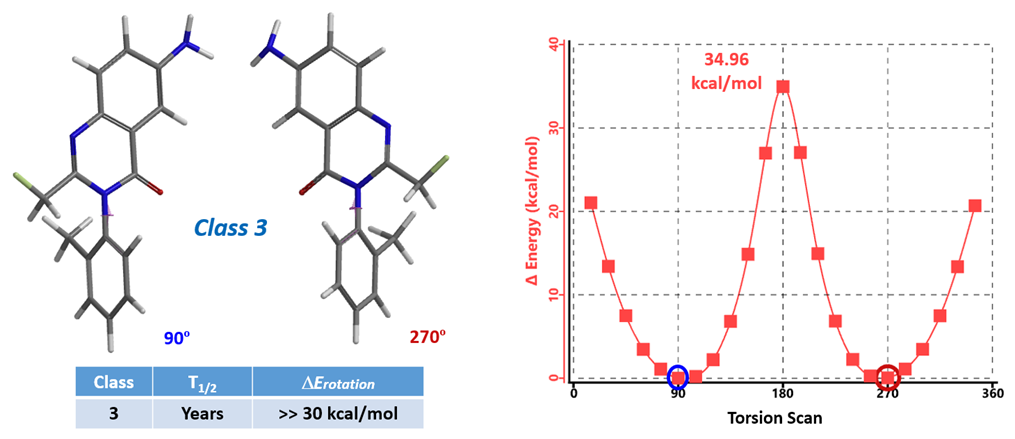
Figure 4. Energy profile from torsion scan of Afloqualone
Drug design can be guided by calculating the interconversion energy barriers of atropisomers, ahead of synthesis. If targeted molecules are predicted to belong to class 2, one could consider increasing the steric hindrance to increase the interconversion energy barrier and transform the compound to class 3 or remove some groups to reduce the steric hindrance and convert it to class 1. And the potential option of eliminating atropisomerism altogether by using the same substituents to make the molecules symmetrical.
Atropisomers could have different conversion rates in different environments. Atropisomers of HIV-1 integrase inhibitor MK-0536 [5] and endothelin type A antagonist BMS-207940 [6] have been reported to have accelerated interconversion in plasma/serum (Figure 5).
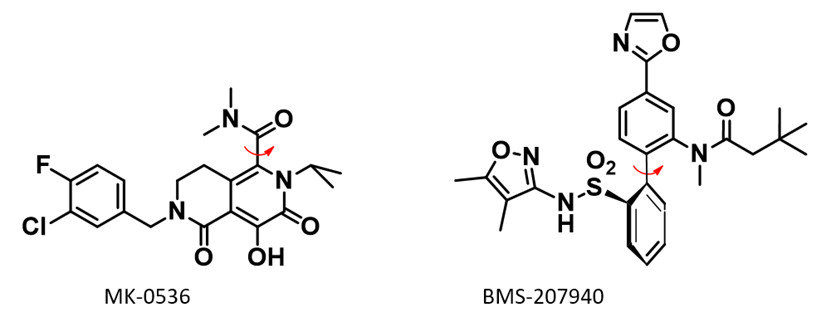
Figure 5. Molecular structure of MK-0536 and BMS-207940
MK-0536 atropisomerism comes from restricted rotation of the C-C bond marked with the red arrow (Figure 5). QM torsion scan shows that when the two lowest conformations have dihedral angle of 90º and 270º, respectively, the energy barrier for their interconversion is 27.29 kcal/mol (Figure 6, the N-benzyl group is simplified to N-methyl group for calculation). As such, MK-0536 belongs to class 2 atropisomer. Under normal conditions, half-life of interconversion will be in days.
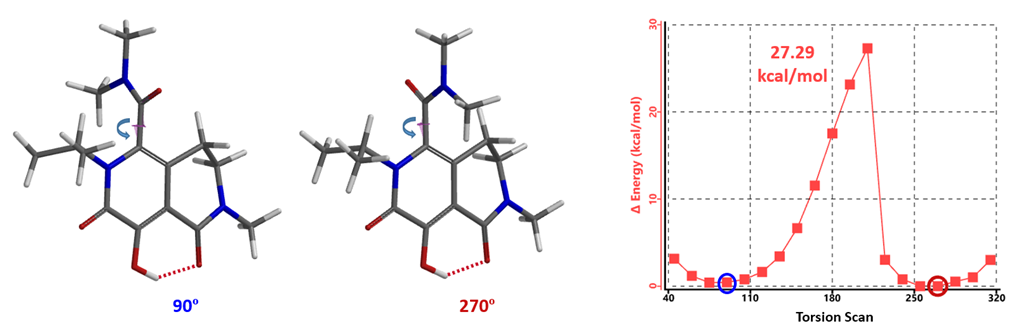
Figure 6. Torsion scan energy profile of MK-0536
However, when MK-0536 was incubated in human plasma at a concentration of 9 mM, the interconversion was completed in <30 minutes, and the T1/2 of the racemization is too fast to be measured by standard methods [5]. The rapid interconversion was also observed in dog, rat, and rhesus monkey plasma. With the rapid rate of interconversion in plasma, separation of the atropisomers is not necessary for clinical development of MK-0536.
The above observations remind us that for class 2 atropisomers, it is prudent that we shall determine their interconversion rate in plasma/serum to make informed decisions for their development strategies.
In this chapter, we discussed the use of QM torsion scan to calculate for energy barrier for interconversion of atropisomers, to guide medicinal chemists to design of drug molecules and establish development strategies. We also learned that plasma/serum could accelerate interconversion of class 2 atropisomers.
Building on What We Just Learned
Shown in Figure 7 is the asymmetrical energy profile obtained from torsion scan of compound A. Why is it asymmetrical? Based on the calculated energy barriers, shall we classify it as class 2 or 1? Is chiral SFC purification necessary?
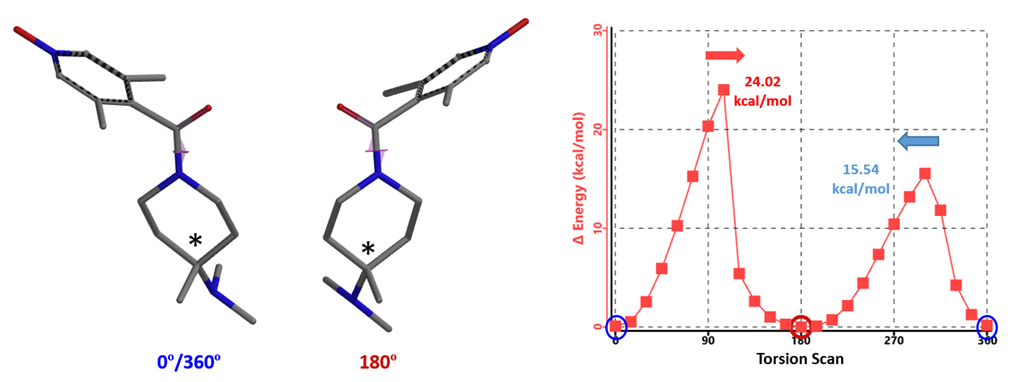
Figure 7. Torsion scan energy profile of compound A
This article is written and edited by Haowei Wang, Xiaoli Shen, Dong Pan, Tommy Lai, Yongsheng Chen, John S. Wai
References:
[1] https://cen.acs.org/pharmaceuticals/drug-development/Giving-atropisomers-another-chance/96/i33.
[2] S.T. Toenjes, J.L. Gustafson, Future Med. Chem. 2018, 10, 409. doi.org/10.4155/fmc-2017-0152.
[3] S.R. LaPlante, L.D. Fader, K.R. Fandrick, D.R. Fandrick, O. Hucke, R. Kemper, S.P.F. Miller, P.J. Edwards, J. Med. Chem. 2011, 54, 7005. S.R. LaPlante, P.J. Edwards, L.D. Fader, A. Jakalian, O. Hucke, ChemMedChem 2011, 6, 505.
[4] J. Chandrasekhar, R. Dick, J. Van Veldhuizen, D. Koditek, E-I. Lepist, M.E. McGrath, L. Patel, G. Phillips, K. Sedillo, J. R. Somoza, J. Therrien, N. A. Till, J. Treiberg, A. G. Villaseñor, Y. Zherebina, S. Perreault, J. Med. Chem. 2018, 61, 6858. L. Xing, B. Devadas, R. V. Devraj, S. R. Selness, H. Shieh, J. K. Walker, M. Mao, D. Messing, B. Samas, J. Z. Yang, G. D. Anderson, E. G. Webb, J. B. Monahan, ChemMedChem 2012, 7, 273.
[5] M.S. Egbertson, J.S. Wai, M. Cameron, R.S. Hoerrner. Discovery of MK-0536: A Potential Second-Generation HIV-1 Integrase Strand Transfer Inhibitor with a High Barrier to Mutation in Antiviral Drugs: From Basic Discovery through Clinical Trials, W.M. Kazmierski, Ed.; Wiley: Hoboken, New Jersey, 2011, Chapter 12, pp 167-8.
[6] Y. Shi, M.H. Huang, J.E. Macorb, D.E. Hughes, J. Chromatogr. A, 2005, 67, 1078.