 Choose language
Choose language
< Return to main menu
 Choose language
Choose language

Antibody-Drug Conjugate (ADC) is complex molecule composed of biologically active small molecule linked to an antibody (mAb) through a cleavable linker. The mAb acts as a specific carrier to transport the small molecule into the target cells. Linkers are usually based on chemical motifs such as hydrazones, peptides, and disulfides. In this chapter, we will discuss the optimization of the disulfide linker for ADC drug discovery.
Conjugation makes use of the sulfhydryl group of cysteines (mAb-SH) on monoclonal antibody to undergo a SN2 type reaction with the disulfide bond on the linker [1] to generate the target ADC (Figure 1, product A). Specific antibody is chosen for target delivery of the ADC into tumor cells. Glutathione, which is high in concentration in tumor cells, will reduce the disulfide bonds, release the highly active small molecule drugs for cell kill.
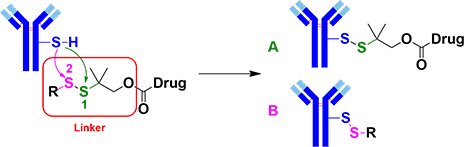
Figure 1. Preparation of ADC by thiol-disulfide exchange reaction
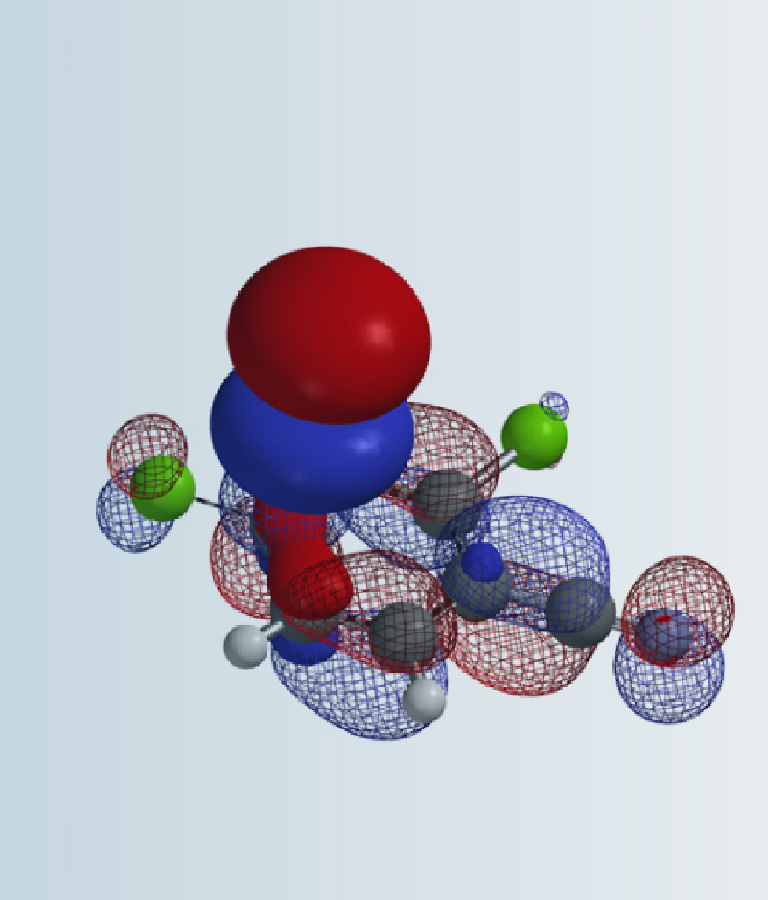
To improve ADC in vivo stability, two methyl groups are usually added at the α position of the central sulfur atom S1 [2] to minimize undesired dissociation of ADC before it reaches the target cells. However, this modification leads to mAb-SH attacking at the undesired S2 during conjugation, resulting in by-product B (Figure 1).
As such, we need to optimize the Linker for selective generation of product A, improving the Drug/Antibody Ratio (DAR) value of ADC target.
In our case, the antibody contains two free cysteine thiols (mAb-SH), giving a maximum DAR value of 2, which is our design goal so as to make full use of it for each mAb internalized into the cancer cells.
General organic chemistry taught us that the lower the pKa value of the conjugated acid of the leaving group R-S- (Figure 1), the better it is as leaving group. Then in principle, conjugation with linker-drugs derived therefrom would provide more target products A, i.e., a higher DAR value. However, such correlation was not observed in our results (Figure 2).
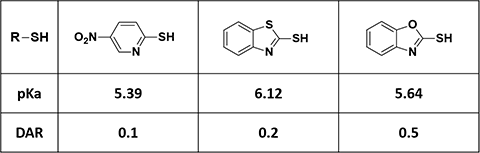
Figure 2. pKa of conjugated acids (leaving group) and corresponding DAR values [3]
We decided to analyze the above results with Quantum Mechanics.
For thiol exchange to proceed, HOMO of mAb-SH needs to interact with LOMO on either S1 or S2. Selectivity will depend on relative size of the LUMO lobes. Shown on Figure 3 are LUMO of the three disulfide linkers above. LUMO lobe on the sulfur atom S1 gradually increases from left to right, and the corresponding DAR value improves accordingly (Figure 3). This could be a useful QM parameter to guide linker design.
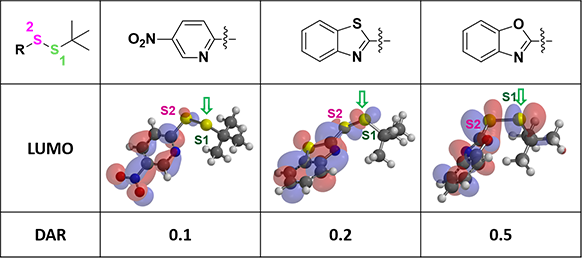
Figure 3. LUMO of linkers and corresponding DAR values
Based on the above analysis, we designed a series of linkers, and evaluated the relative size of their LUMO lobes on sulfur atoms S1 and S2. Isothiourea and methyl sulfone (MTS) derived linkers were then identified with extensive LUMO lobes over the desired S1 (Figure 4). These suggest that mAb-SH could attack selectively at S1 and provide excellent conjugation DAR. To avoid the production of toxic thiourea, we focused our exploration efforts on MTS.
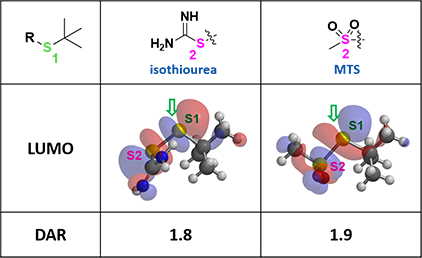
Figure 4. LUMO of isothiourea and MTS linkers and corresponding DAR values
Even though synthesis of the key linker for conjugation with mAb-SH encountered significant difficulties, LUMO analysis provides us with compelling reasons to persist and prepare the MTS linker-drug. A DAR of 1.9 was obtained in conjugation!
In summary, we established the use of LUMO to guide the design of disulfide linkers, discovered MTS linker, and improved DAR by nearly 20 folds over the nitropyridine. A major hurdle for mAb thiol exchange is resolved [4].
In the next chapter, we will discuss the application of QM to analyze Torsional Control in stereo-selective reaction. Please stay tuned.
This article is written and edited by Zijin Xu, Jinhua Chen, Dong Pan, Guqin Shi, and John S. Wai.
References:
[1] Robert D. Bach, Olga Dmitrenko, and Colin Thorpe., J. Org. Chem., 2008, 73, 12.
[2] Ravi V. J. Chari, Michael L. Miller, and Wayne C. Widdison., Angew. Chem. Int. Ed., 2014, 53, 3796.
[3] Application of Electrostatic Potential Map in Estimation of Acidity http://rsd.wuxiapptec.com/qm-magic-class_chapter-3
[4] Jack D. Sadowsky, Thomas H. Pillow, Jinhua Chen, Hui Yao, Zijin Xu, and John Wai, et al., Bioconjugate Chem., 2017, 28, 2086.