 Choose language
Choose language
< Return to main menu
 Choose language
Choose language

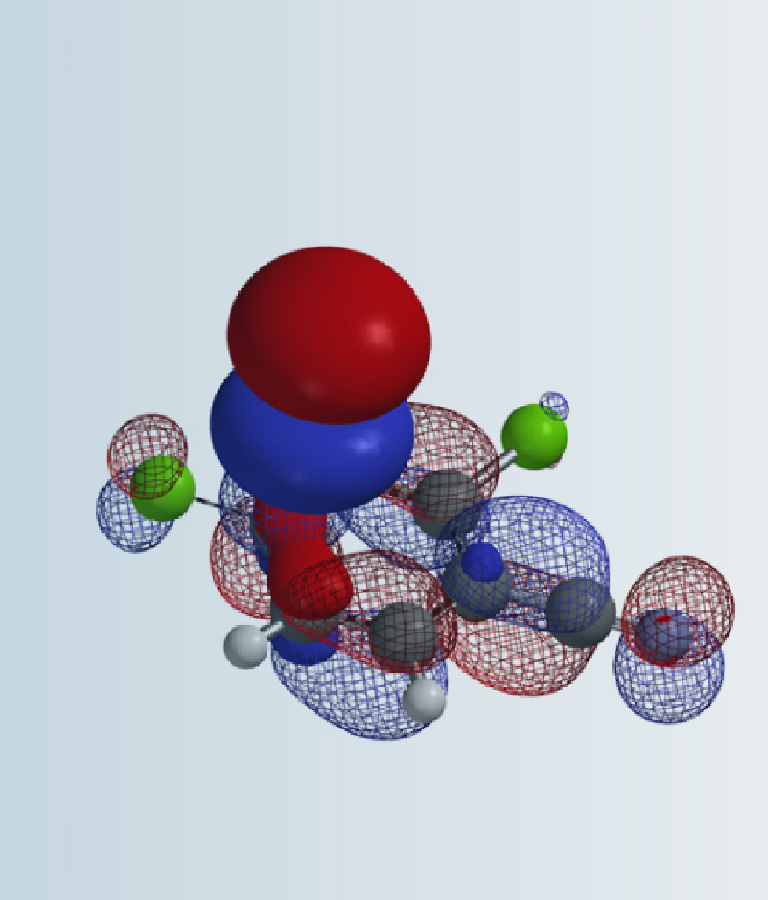
In organic synthesis, for reactions that produce a mixture of isomeric products, we usually need to use various 2D NMR spectroscopic methods to assign their structures. Yet, there are still many instances that we could not do so conclusively.
In this chapter, we will discuss the use of Quantum Mechanics calculated 13C NMR data to assist structural assignment of these compounds.
From a Chan-Lam coupling reaction of a chloropyrazolopyridine, we obtained a mixture of regio-isomers (Figure 1). 2D NMR spectroscopy did not provide us with the necessary C-H or H-H correlation to differentiate them conclusively.
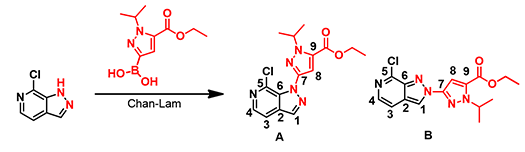
Figure 1. Reaction products from Chan-Lam reaction with pyrazolopyridine
We did 13C NMR calculations for structures A and B with Spartan software [1] and compared with the spectroscopic data obtained experimentally. Tabulated in Table 1 are the experimental 13C NMR data of product 1 and the calculated values for structures A and B. Based on the differences, product 1 is more likely to have the structure A.
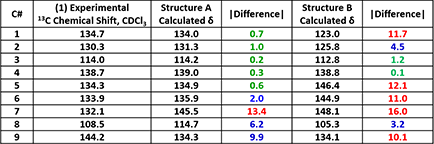
able 1. Comparison of experimental and calculated 13C NMR data between product 1 and structures A and B, respectively (Spartan '16)
Note: Green: Δ < 2 ppm, blue: 2 ≤ Δ ≤ 10 ppm, red indicates: Δ > 10 ppm. |Difference| represents the absolute value of the difference between the experimental and calculated values. Same for following Tables.
Similarly, we compared the experimental data of product 2 with the calculated values (Table 2). Product 2 aligned with structure B much better.

Table 2. Comparison of experimental and calculated 13C NMR data between product 2 and structures A and B, respectively (Spartan '16)
This allows us to tentatively assign the structures to product 1 and 2 with confidence. Subsequent experimental results substantiate the assignments.
We were curious to know whether this could be applied to natural products with more complex structures. Below is one of the examples we looked at.
In 2010, Yongsheng Che et al. reported the discovery of Cytosporolides A - C with antibacterial activity in Org. Lett. [2]. Proposed structure for Cytosporolides A and its absolute stereochemistry assignment were based on 2D NMR spectroscopy, single crystal X-ray diffraction, data in the literature, etc.
The antibacterial properties of Cytosporolides and their unique structures attracted significant interest among synthetic chemists. In 2011, Australian scientists Jonathan H. George et al., based on chemical synthesis and data analysis, proposed an alternate six-membered aryl ether structure for Cytosporolides A (Figure 2), and published their results in Org. Lett. [3].
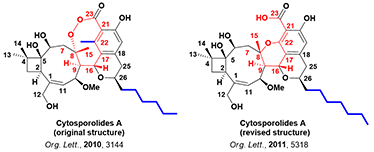
Figure 2. Proposed structures for Cytosporolides A
The key structural differences of the two proposed structures are highlighted in red and blue (Figure 2). Calculated 13C NMR data of the proposed structures for the key signals for C7-C9, C17-C23, and C25 were compared with experimental ones (Table 3). The data are supportive of the structure proposed by George et al.
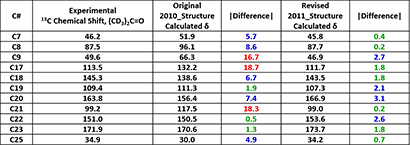
Table 3. Comparison of experimental and QM-calculated 13C NMR data with the two proposed structures of Cytosporolides A. Tabulated are the core differences. (Spartan '18)
The above cases highlighted the application of QM NMR calculation to assist structure assignments, from relatively simple structures to more complicated natural products. In our laboratories, we set up the NMR experiments and calculations concurrently to enable proper structure assignments in a timely manner.
NMR algorithm in the Spartan software was optimized with experimental data obtained with CDCl3 as solvent. It is prudent to obtain experimental data in CDCl3 for proper comparison.
This article is written and edited by Dajin Tan, Yongsheng Chen, Guqin Shi, Qiuyue Wang, and John S. Wai.
References
[1] a. Warren J. Hehre (2019). Spartan '18 Tutorial and User's Guide, Calculating NMR Spectra, page 525-529, Irvine, CA, USA: Wavefunction, Inc. b. Warren Hehre, J. Nat .Prod., 2019, 82, 2299.
[2] Yongsheng Che, et al., Org. Lett., 2010, 12, 3144.
[3] Jonathan H. George, et al., Org. Lett., 2011, 13, 5318.